[,1] [,2]
[1,] 50.3 -49.7
[2,] -49.7 50.3Ridge Regression and Cross Validation
MSSC 6250 Statistical Machine Learning
Department of Mathematical and Statistical Sciences
Marquette University
Ridge Regression
When Ordinary Least Squares (OLS) Doesn’t Work Well: Collinearity
- When predictors are highly correlated, \(\mathrm{Var}(b_j)\) is much inflated.1
- A tiny change in the training set causes a large change in \(b_j\), leading to unreliable estimation and possibly prediction.
\({\bf X'X} = \begin{bmatrix} 1 & 0.99 \\ 0.99 & 1 \end{bmatrix}\) \(\quad ({\bf X'X})^{-1} = \begin{bmatrix} 50.3 & -49.7 \\ -49.7 & 50.3 \end{bmatrix}\)
-
\(\mathrm{Var}(b_j) = 50.3\sigma^2\)
- An increase in 50-fold over the ideal case when the two regressors are orthogonal.
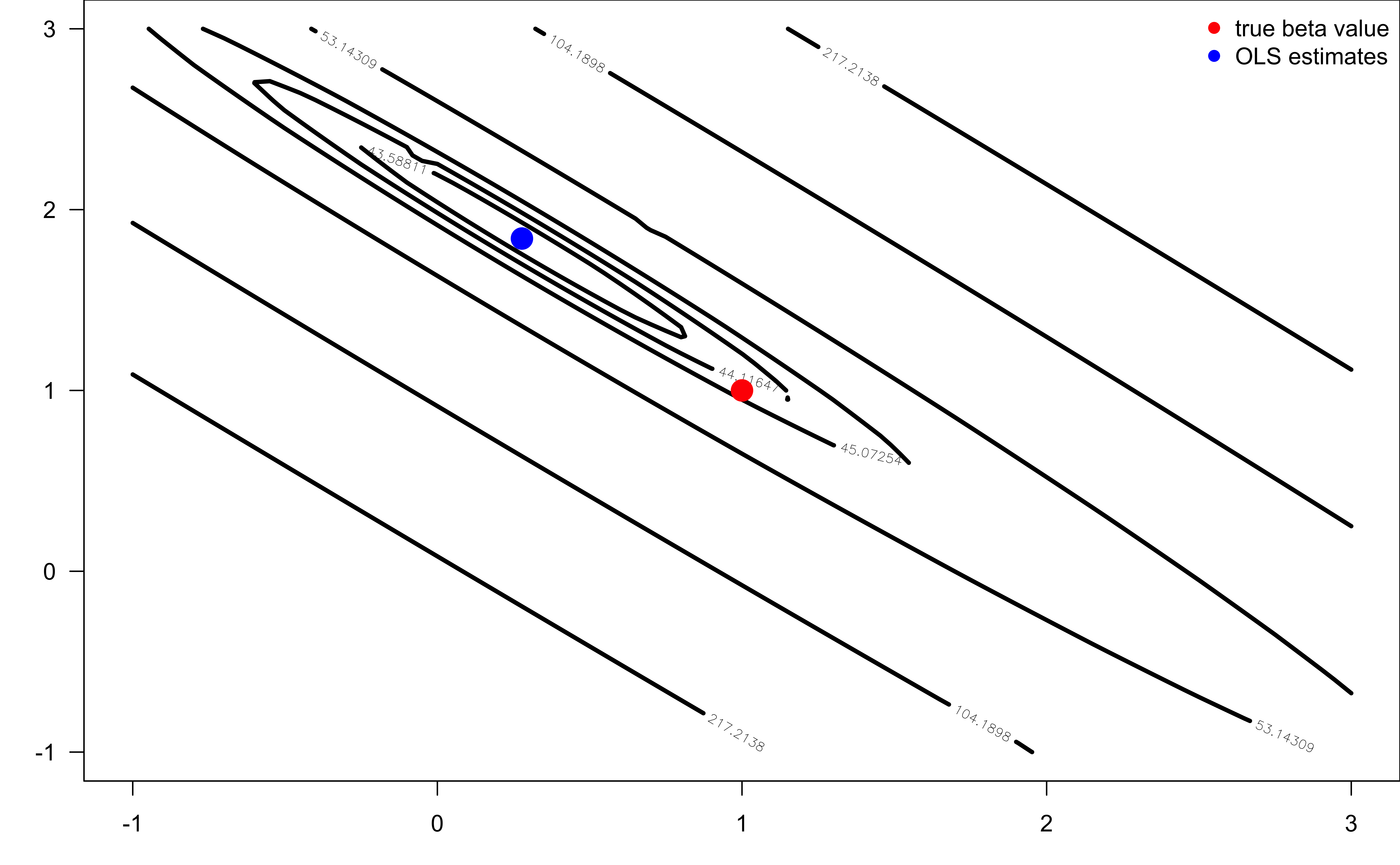
When OLS Doesn’t Work Well: Optimization Perspective
\(\beta_1 = \beta_2 = 1\) and \(\mathrm{Corr}(x_1, x_2) = 0.99\)
The relatively “flat” valley in the objective function walks along the eigen-vector of \({\bf X}'{\bf X}\) having the smallest eigen-value.
Ridge and Valley
- Statisticians used to maximize the likelihood, leading to a “ridge”.
- Computer scientists like to minimize the loss, leading to a “valley”.
When OLS Doesn’t Work Well: Optimization Perspective
- A little change in the training set perturbs the objective function. The LSEs lie on a valley centered around the truth.
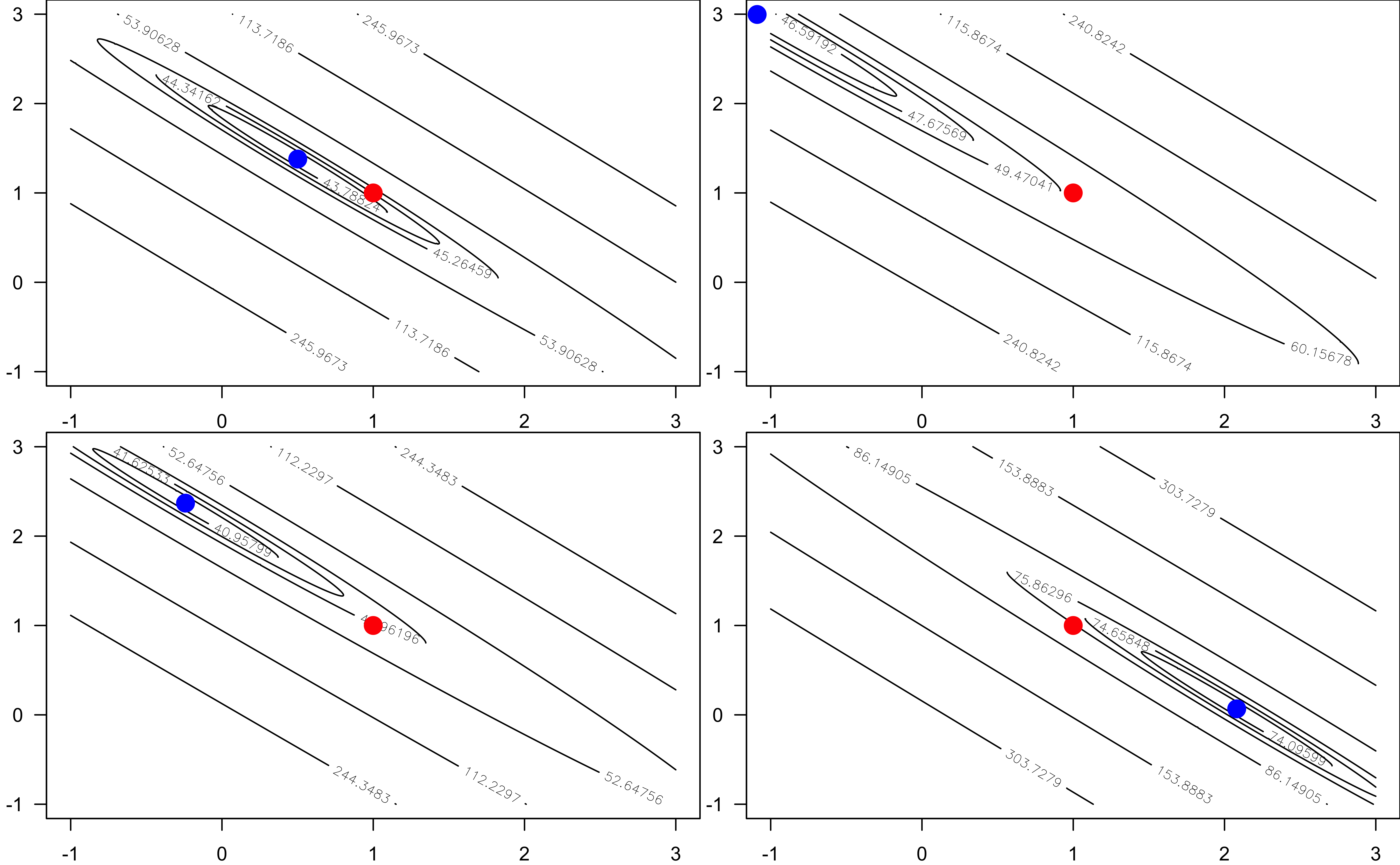
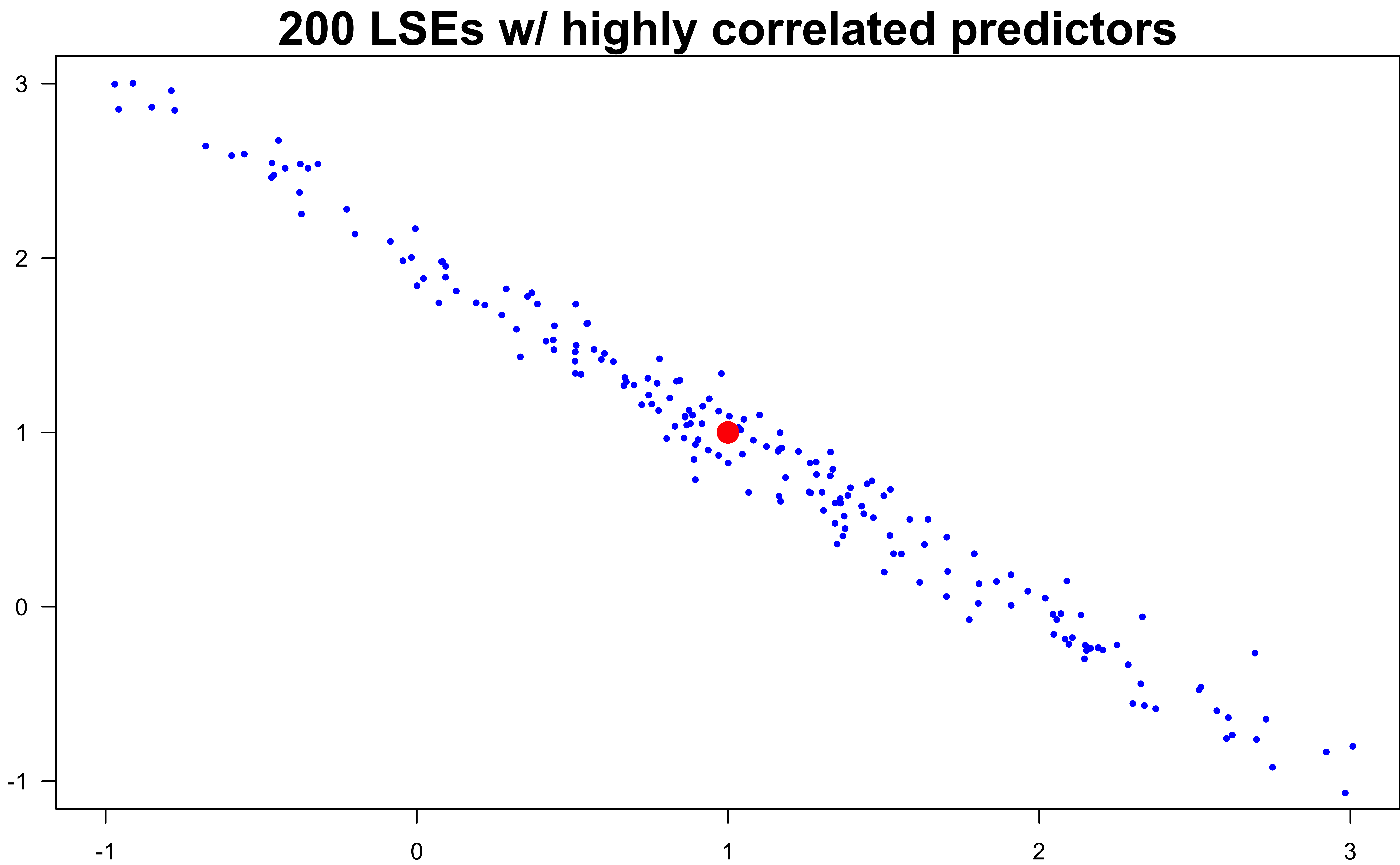
When OLS Doesn’t Work Well: High Variance
The optimizer could land anywhere along the valley, leading to large variance of the LSE.
Over many simulation runs, the LSE lies around the line of \(\beta_1 + \beta_2 = 2\), the direction of the eigen-vector of the smallest eigen-value.
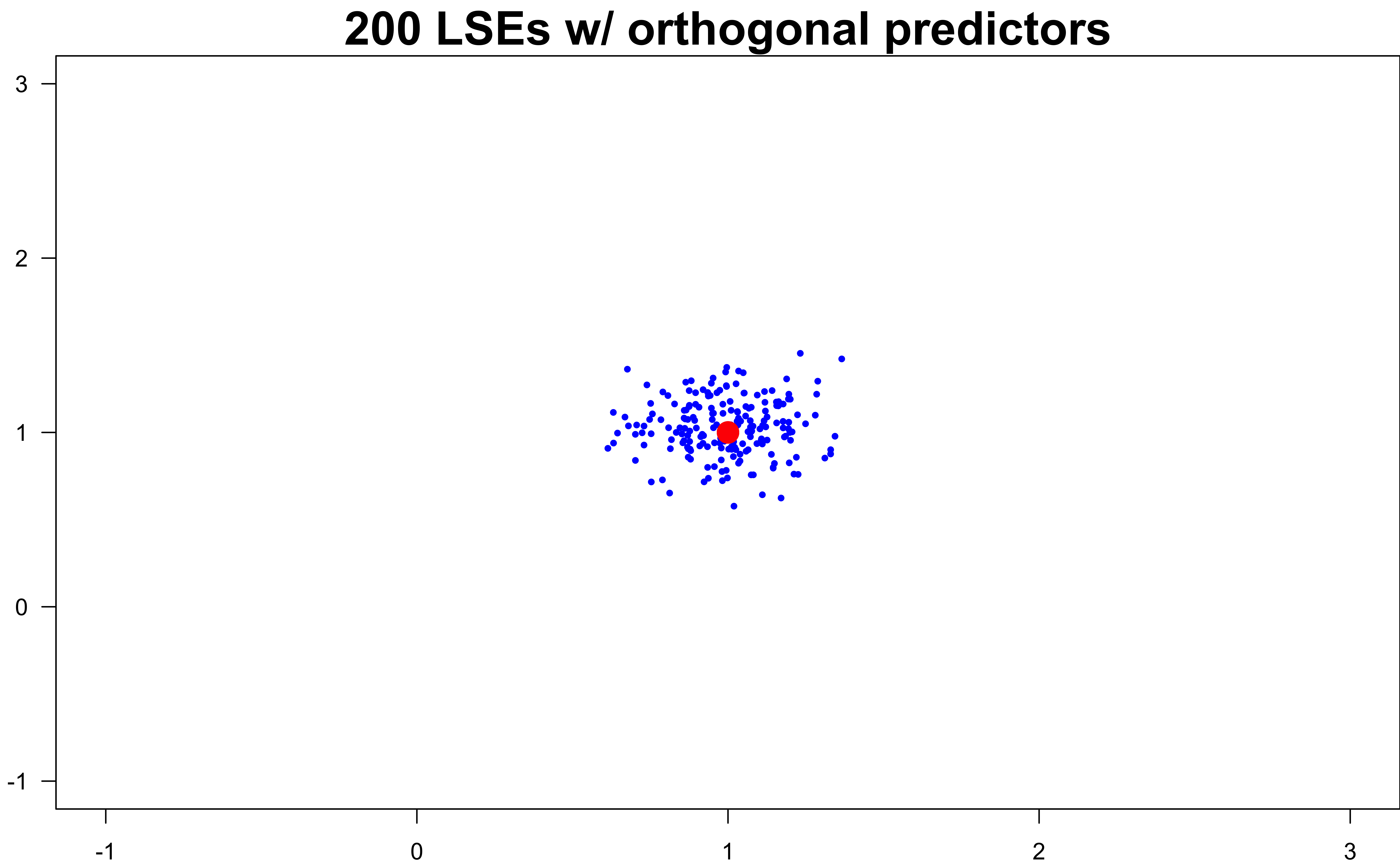
When OLS Doesn’t Work Well: Large-\(p\)-small-\(n\) (High Dimensional Data)
OLS stays well in the world of “large-\(n\)-small-\(p\)”.
When \(p > n\), \({\bf X}'{\bf X}\) is not invertible.
There is no unique \(\boldsymbol \beta\) estimate.
Intuition: Too many degrees of freedom (\(p\)) to specify a model, but not enough information (\(n\)) to decide which one is the one.
- Too flexible and ends up with overfitting
Remedy for Large Variance and Large-\(p\)-small-\(n\)
Make \({\bf X}'{\bf X}\) invertible when \(p > n\) by regularizing coefficient behavior!
A good estimator balances bias and variance well, or minimizes the mean square error \[\text{MSE}(\hat{\beta}) = E[(\hat{\beta} - \beta)^2] = \mathrm{Var}(\hat{\beta}) + \text{Bias}(\hat{\beta})^2\]
A slightly biased estimator \(\hat{\boldsymbol \beta}\) that has much smaller variance and MSE than the LSE \({\bf b}\).
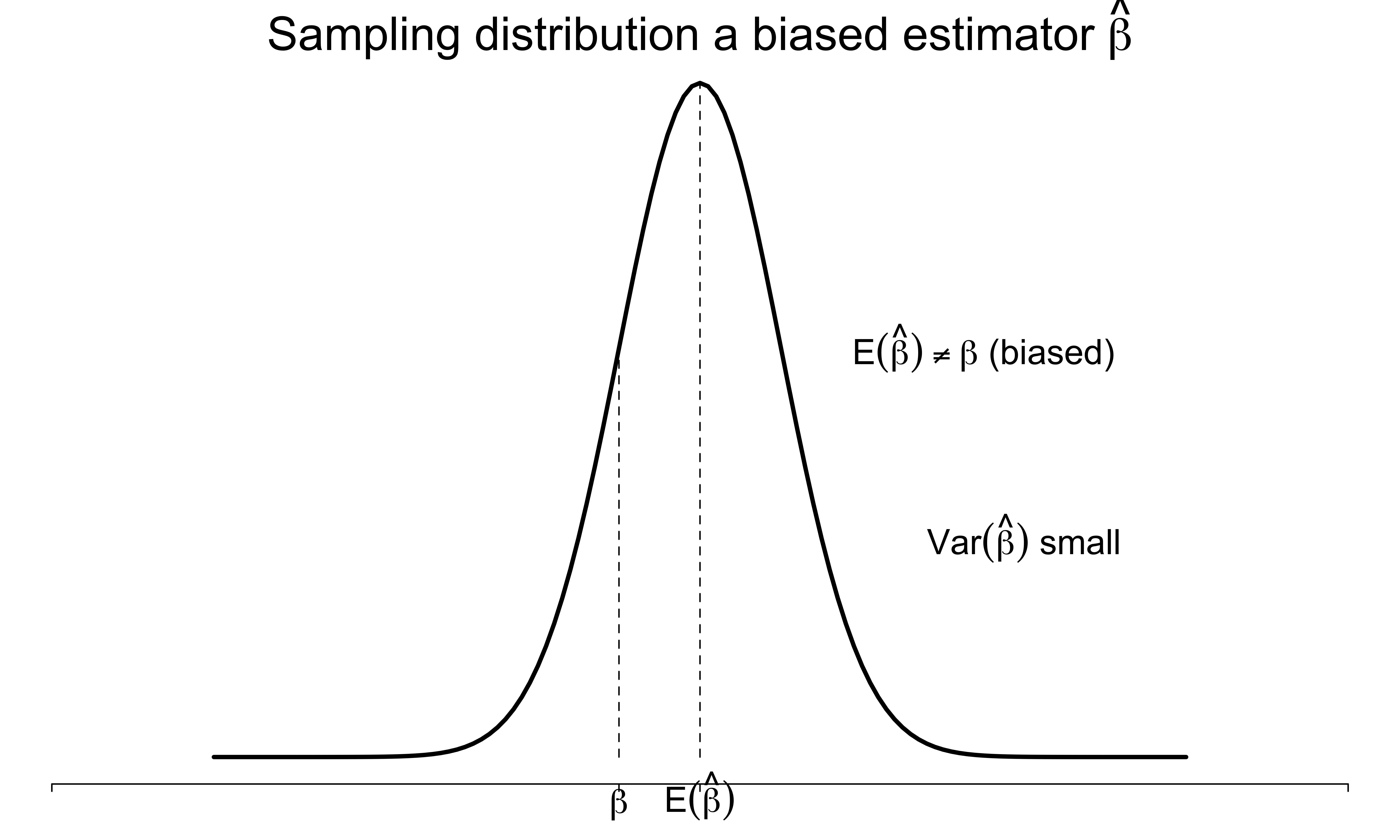
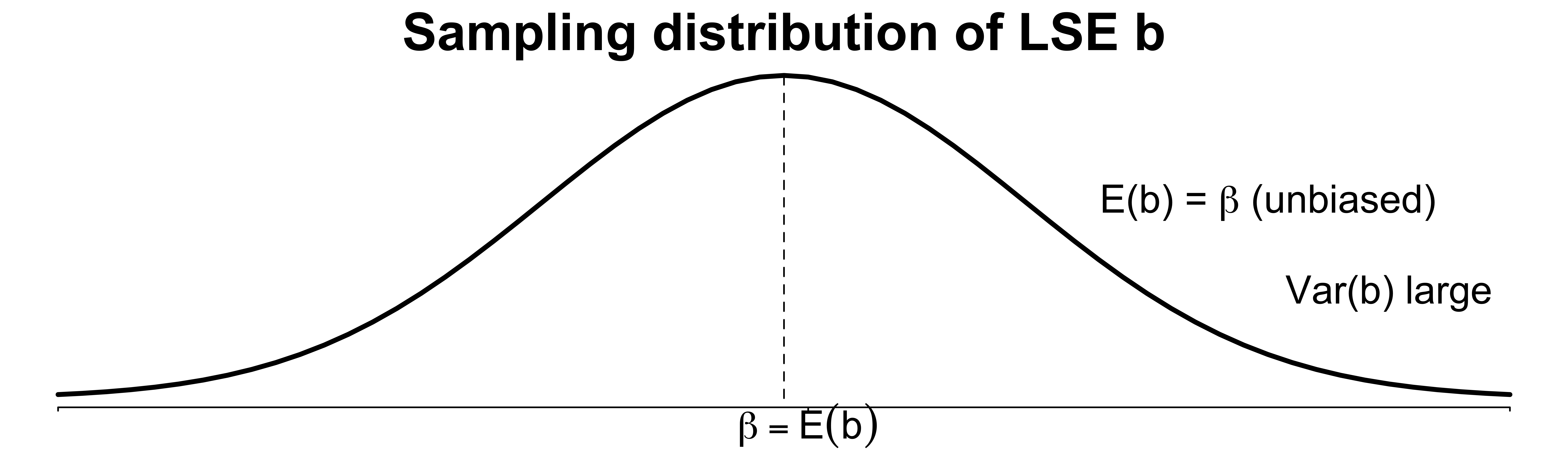
Ridge Regression
- Idea: Add a ridge (diagonal matrix) to \({\bf X} ' {\bf X}\).1 \[\widehat{\boldsymbol \beta}^\text{r} = (\mathbf{X}' \mathbf{X}+ n \lambda \mathbf{I})^{-1} \mathbf{X}' \mathbf{y},\]
- To regularize coefficients, add an \(\ell_2\) penalty to the residual sum of squares, for some tuning parameter \(\lambda > 0\).
\[ \begin{align} \widehat{\boldsymbol \beta}^\text{r} =& \, \mathop{\mathrm{arg\,min}}_{\boldsymbol \beta} \underbrace{\lVert \mathbf{y}- \mathbf{X}\boldsymbol \beta\rVert_2^2}_{SS_{res}} + n \lambda \lVert\boldsymbol \beta\rVert_2^2\\ =& \, \mathop{\mathrm{arg\,min}}_{\boldsymbol \beta} \underbrace{\frac{1}{n} \sum_{i=1}^n (y_i - x_i'\boldsymbol \beta)^2}_{\text{MSE}_{\texttt{Tr}}} + \lambda \sum_{j=1}^p \beta_j^2\\ =& \, \mathop{\mathrm{arg\,min}}_{\boldsymbol \beta} \color{green} - \text{ goodness of fit } + \text{ model complexity/flexibility} \end{align} \]
Properties of Ridge Regression
\[ \begin{align} \widehat{\boldsymbol \beta}^\text{r} =& \mathop{\mathrm{arg\,min}}_{\boldsymbol \beta} \frac{1}{n} \sum_{i=1}^n (y_i - x_i'\boldsymbol \beta)^2 + \lambda \sum_{j=1}^p \beta_j^2, \end{align} \]
Properties of ridge regression:
- Has less degrees of freedom in the sense that the cost gets higher when larger coefficients are used.
- Favors small-magnitude coefficient estimates (Shrinkage) to avoid cost penalty.
- The shrinkage parameter \(\lambda\) controls the degree of penalty.
- \(\lambda \rightarrow 0\): No penalty, and \(\widehat{\boldsymbol \beta}^\text{r} = \bf b\).
- \(\lambda \rightarrow \infty\): Unbearable penalty, and \(\widehat{\boldsymbol \beta}^\text{r} \rightarrow \mathbf{0}\)
Ridge Penalty
\[\lambda \lVert \boldsymbol \beta\rVert^2_2 = \lambda \boldsymbol \beta' \mathbf{I}\boldsymbol \beta\]
The penalty contour is circle-shaped
Force the objective function to be more convex
A more stable or less varying solution
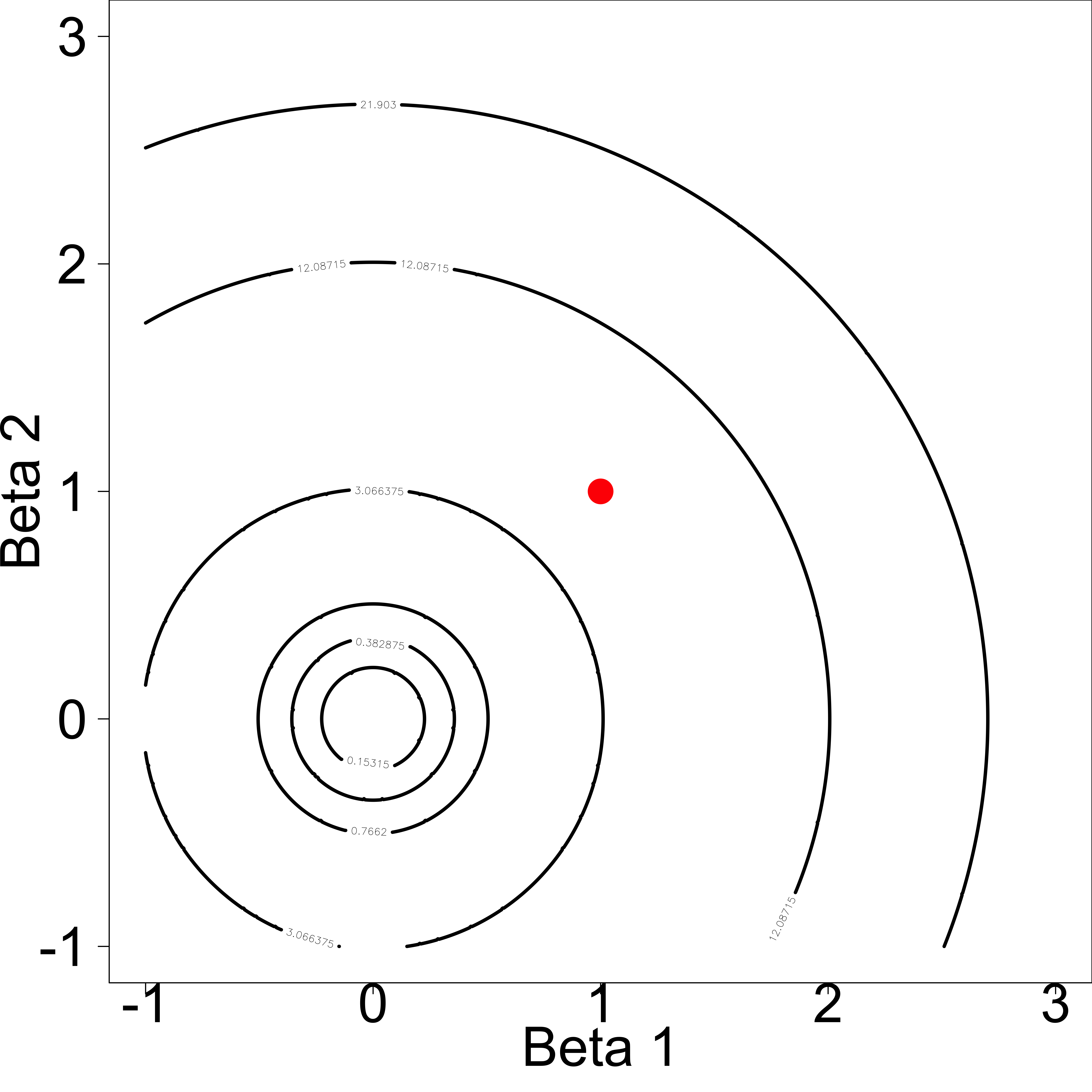
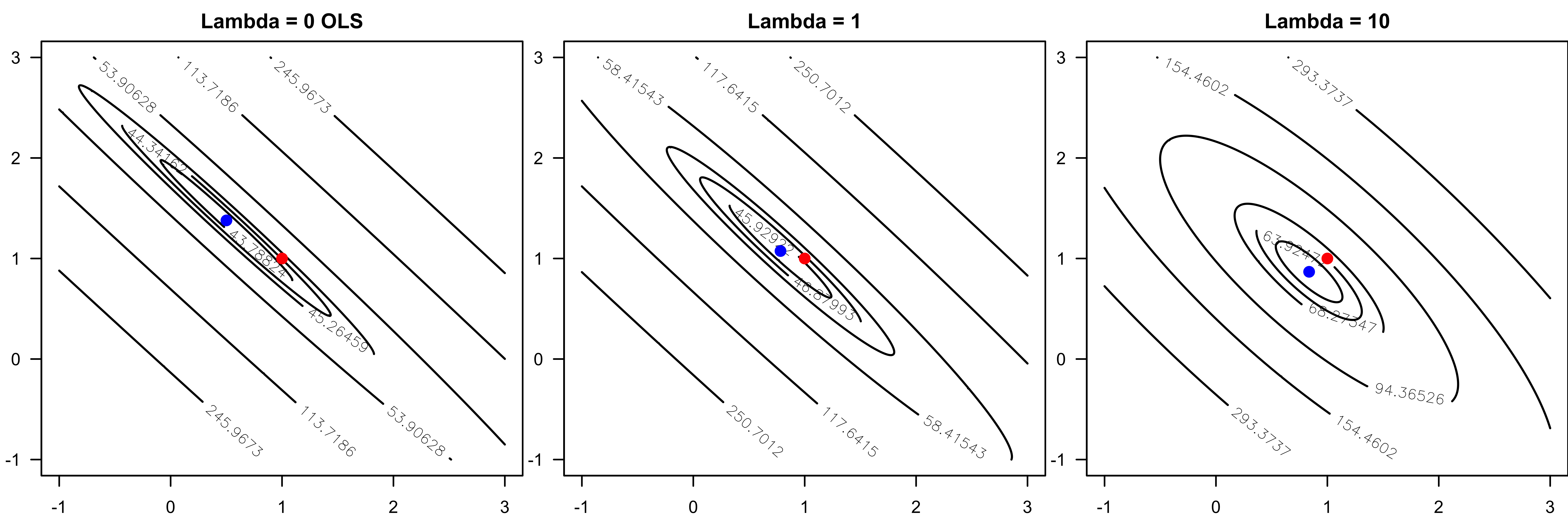
Geometrical Meaning of Ridge Regression
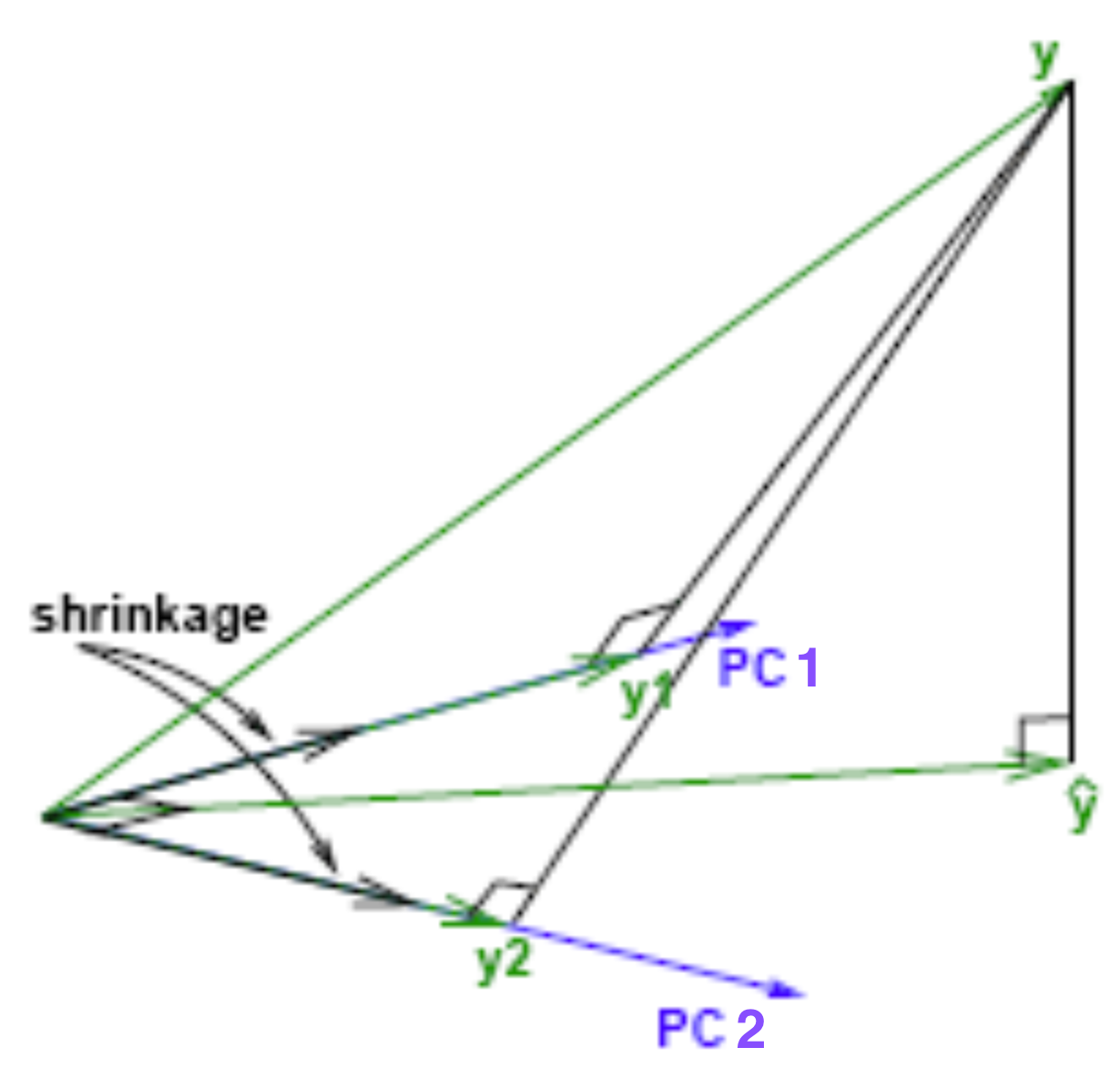
More Convex Loss Function
- Adding a ridge penalty forces the objective to be more convex due to the added eigenvalues.
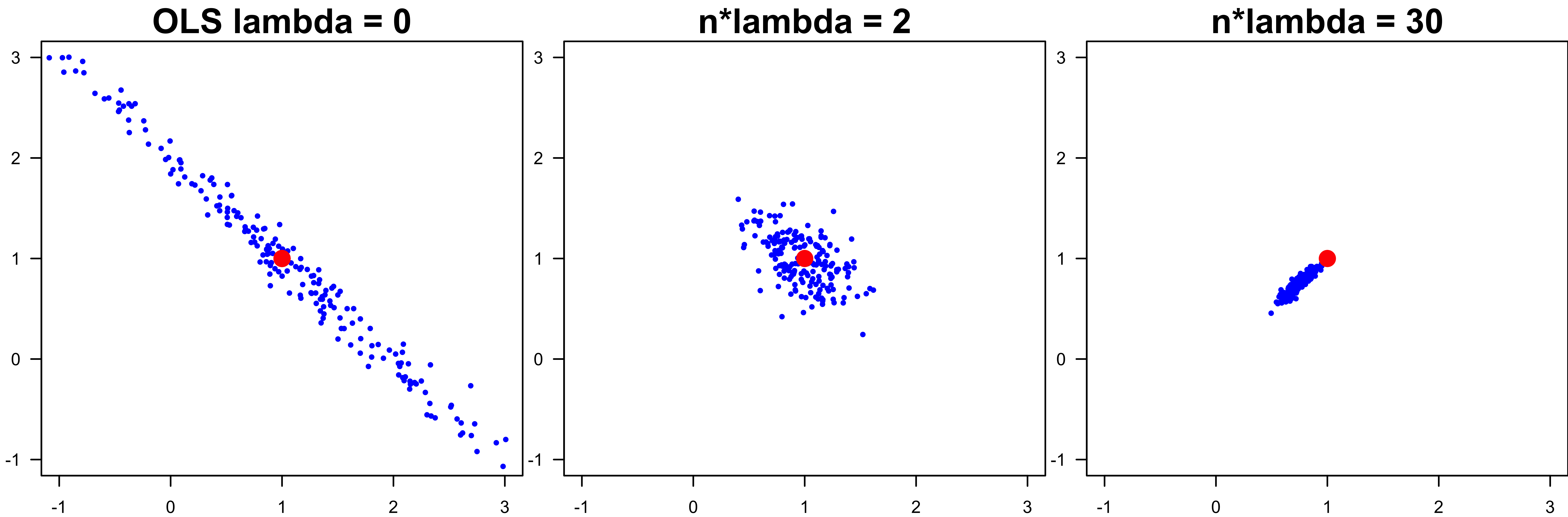
The Bias-variance Tradeoff
- As \(\lambda \downarrow\), bias \(\downarrow\) and variance \(\uparrow\)
- As \(\lambda \uparrow\), bias \(\uparrow\) and variance \(\downarrow\)
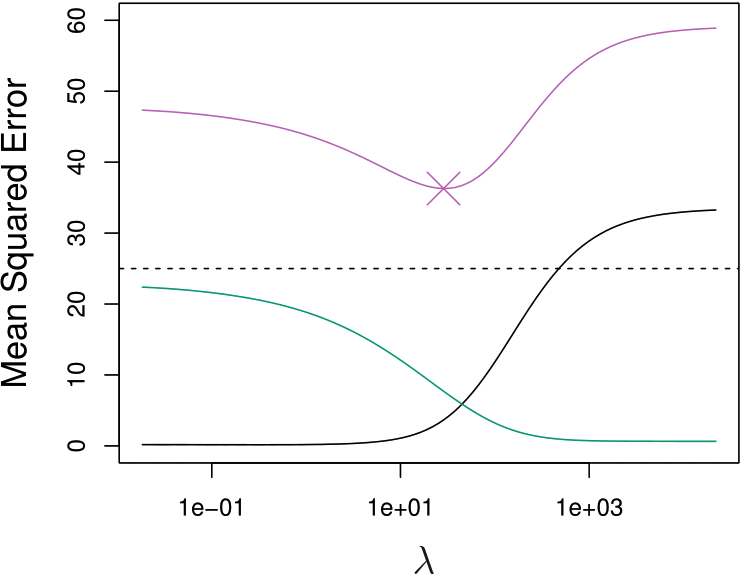
Lower Test MSE
When \(b_j\) has large variance or \(p > n\), ridge regression could have lower test MSE and better predictive performance.
\(\text{MSE}_{\texttt{Tr}}\) (purple)
Squared bias (black)
Variance (green)
MASS::lm.ridge()
The
lambdaparameter inMASS::lm.ridge()specifies the \(n\lambda\) in our notation.OLS is scale equivalent: \(X_jb_j\) remains the same regardless of how \(X_j\) is scaled.
Ridge coefficient estimates can change substantially when multiplying a given predictor by a constant, i.e., \(X_j\hat{\beta}^{r}_{j, \lambda}\) depends on \(\lambda\), the scaling of \(X_j\), and even the scaling of other predictors.
Standardize all predictors!1
head(mtcars, 3) mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.62 16.5 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.88 17.0 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.32 18.6 1 1 4 1(fit <- MASS::lm.ridge(mpg ~ ., data = mtcars, lambda = 1)) ## ridge fit cyl disp hp drat wt qsec vs
16.53766 -0.16240 0.00233 -0.01493 0.92463 -2.46115 0.49259 0.37465
am gear carb
2.30838 0.68572 -0.57579 Scaling of lm.ridge()
coef(fit)transforms these back to the original scale for non-standardized \(\mathbf{X}\).fit$coefshows the coefficients of the standardized predictors.
coef(fit) cyl disp hp drat wt qsec vs
16.53766 -0.16240 0.00233 -0.01493 0.92463 -2.46115 0.49259 0.37465
am gear carb
2.30838 0.68572 -0.57579 fit$coef cyl disp hp drat wt qsec vs am gear carb
-0.285 0.285 -1.008 0.487 -2.370 0.866 0.186 1.134 0.498 -0.915 Ridge Trace
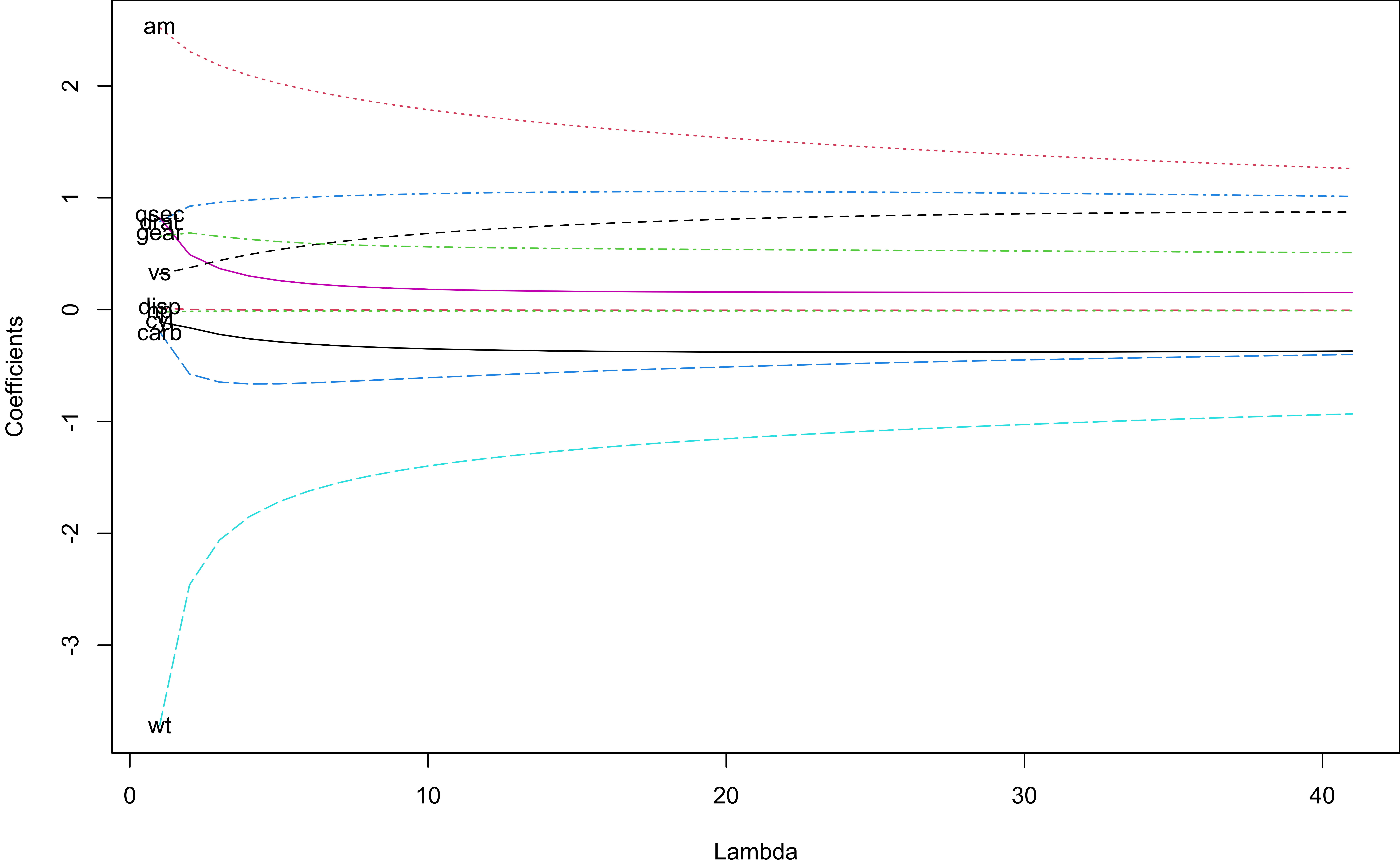
- Select a value of \(\lambda\) at which the ridge estimates are stable.
Methods for Choosing \(\lambda\)
MASS::select(ridge_fit)modified HKB estimator is 2.59
modified L-W estimator is 1.84
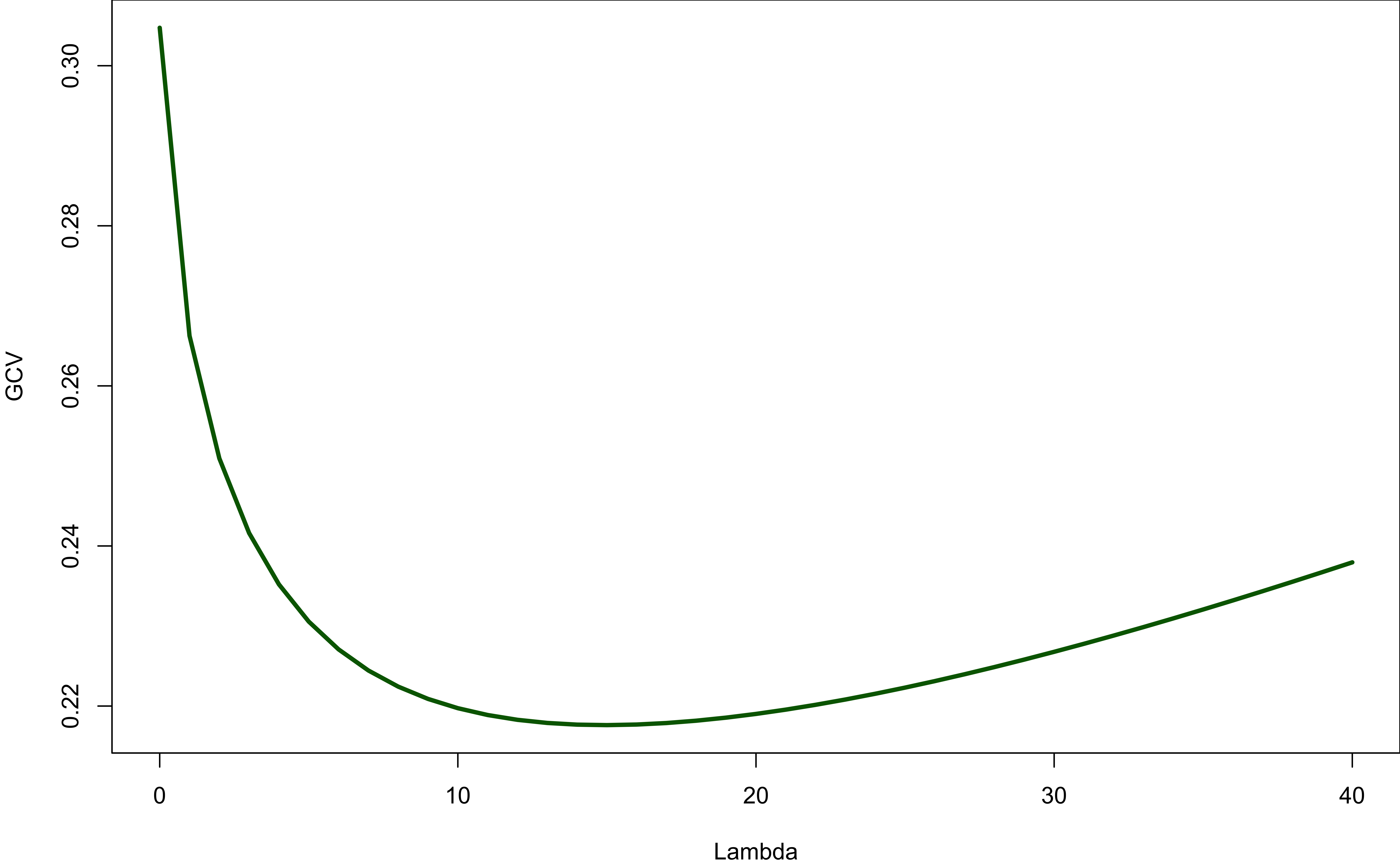
smallest value of GCV at 15 Hoerl, Kennard, and Baldwin (1975): \(\lambda \approx \frac{p \hat{\sigma}^2}{{\bf b}'{\bf b}}\)
Lawless and Wang (1976): \(\lambda \approx \frac{np \hat{\sigma}^2}{{\bf b'X}'{\bf Xb}}\)
Golub, Health, and Wahba (1979): Generalized Cross Validation
Cross Validation
Cross Validation (CV) is a resampling method.
Resampling methods refit a model of interest to data sampled from the training set.
-
CV can be used to
- estimate the test error when there is no test data. (Model assessment)
- select the tuning parameters that controls the model complexity/flexibility. (Model selection)
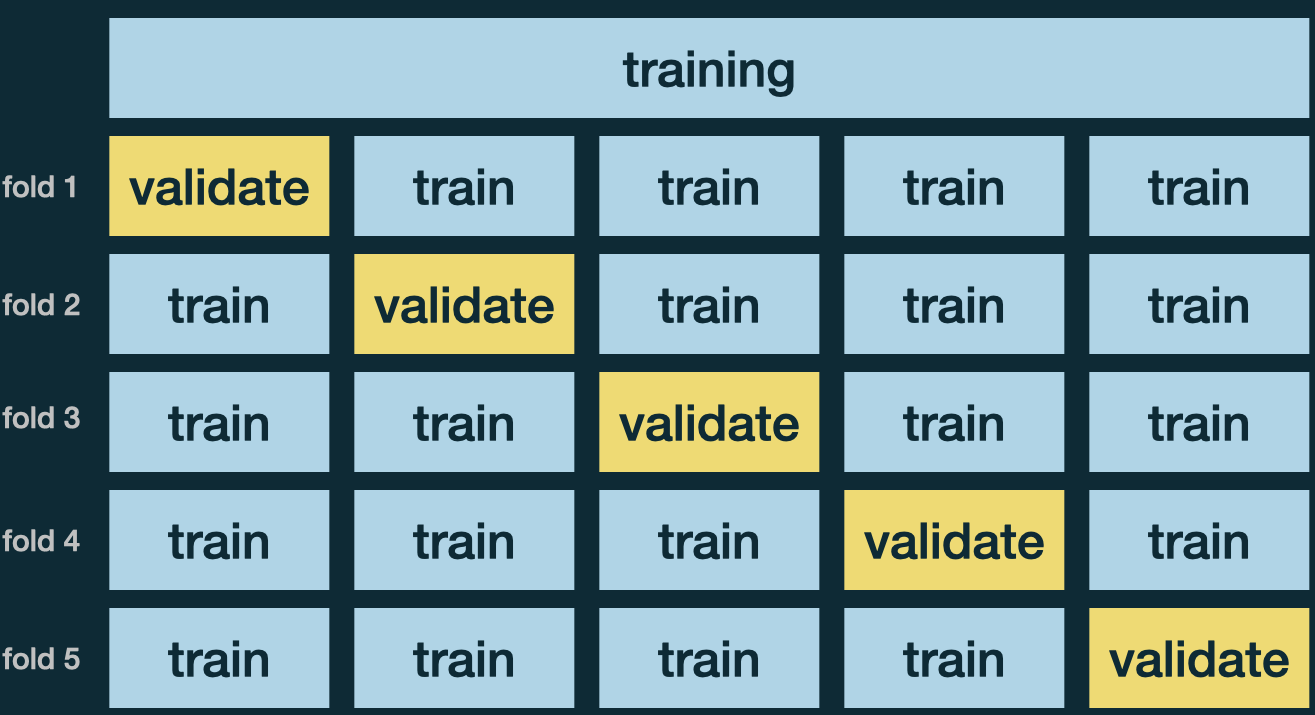
\(k\)-Fold Cross Validation
Randomly divide the training set into \(k\) folds, of approximately equal size.
Use 1 fold for validation to compute MSE, and the remaining \(k - 1\) partitions for training.
Repeat \(k\) times. Each time a different fold is treated as a validation set.
Average \(k\) metrics, \(\text{MSE}_{CV} = \frac{1}{k}\sum_{i=1}^k\text{MSE}_i\).
Use the CV estimate \(\text{MSE}_{CV}\) to select the “best” tuning parameter.
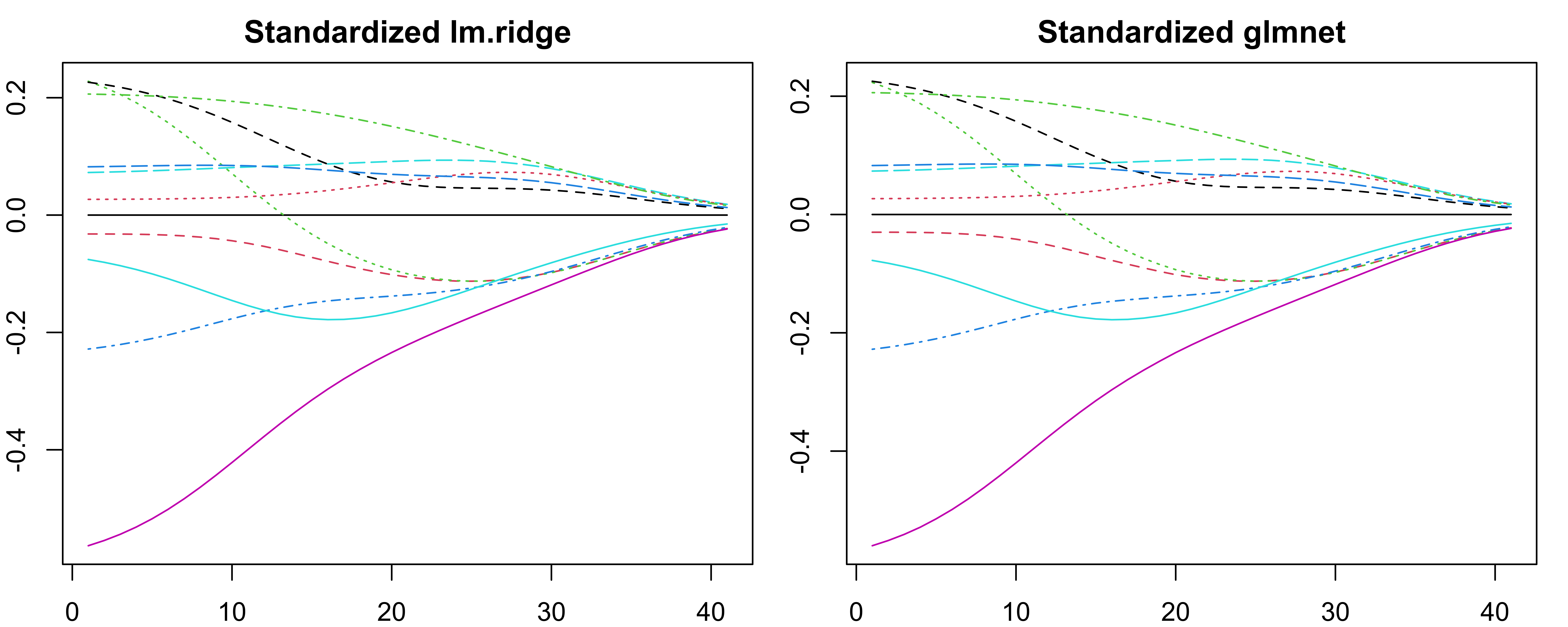
glmnet package ![]()
The parameter
alphacontrols the ridge (alpha = 0) and lasso (alpha = 1) penalties.Supply a decreasing sequence of
lambdavalues.lm.ridge()use \(SS_{res}\) and \(n\lambda\), whileglmnet()use \(\text{MSE}_{\texttt{Tr}}\) and \(\lambda\).Argument
xshould be a matrix.
glmnet(x = data.matrix(mtcars[, -1]),
y = mtcars$mpg,
alpha = 0,
lambda = 5:0/nrow(mtcars))lm.ridge(formula = mpg ~ .,
data = mtcars,
lambda = 5:0)Note
glmnet()only usecoef()and return coefficients at the original scale.lm.ridge()andglmnet()coefficients do not match exactly, especially when transforming back to original scale.No need to worry too much as we focus on predictive performance.

\(k\)-Fold Cross Validation using cv.glmnet()1
- The \(\lambda\) values are automatically selected, on the \(\log_{e}\) scale.
ridge_cv_fit <- cv.glmnet(x = data.matrix(mtcars[, -1]), y = mtcars$mpg, alpha = 0,
nfolds = 10, type.measure = "mse")
plot(ridge_cv_fit$glmnet.fit, "lambda")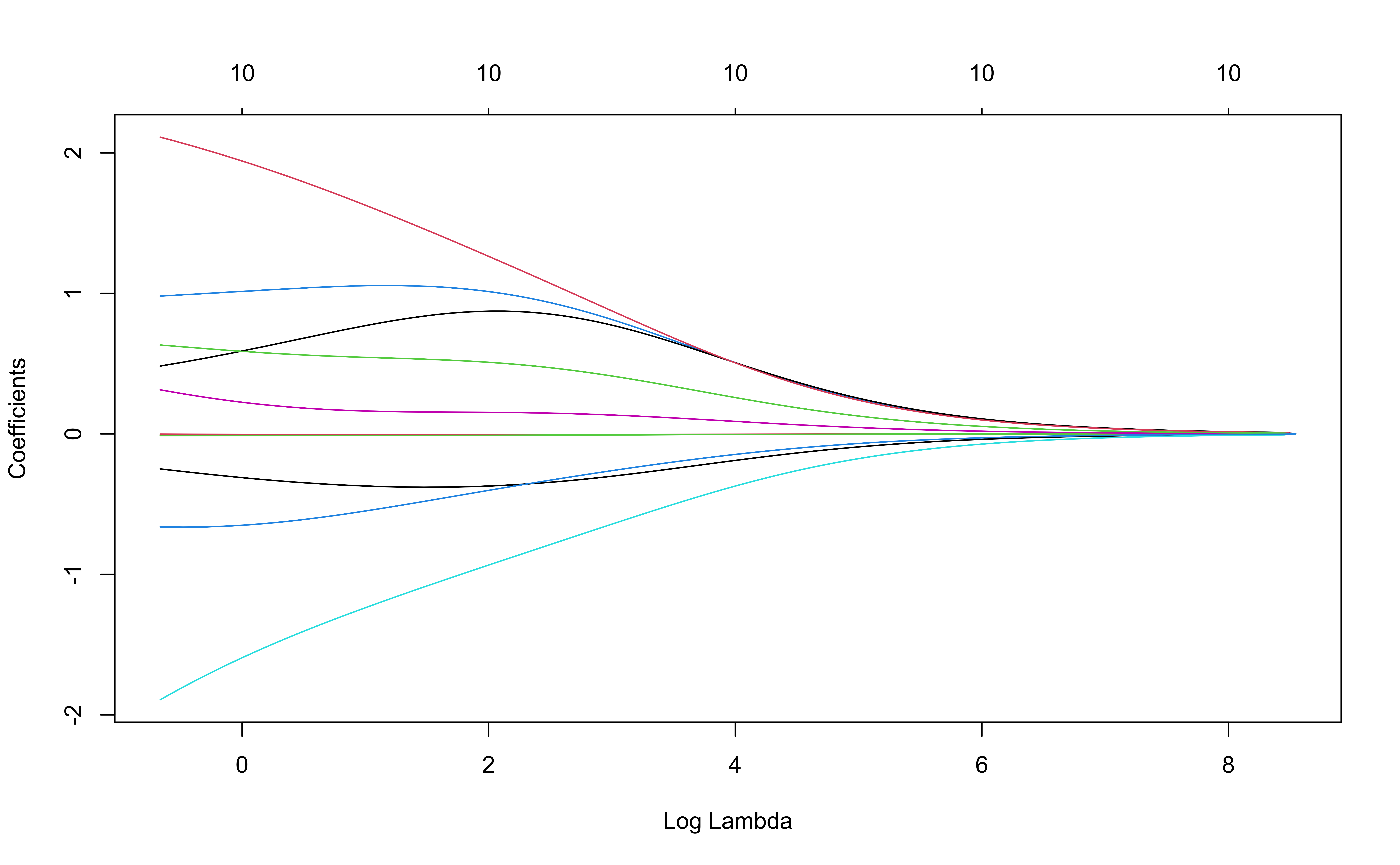
Determine \(\lambda\)
plot(ridge_cv_fit)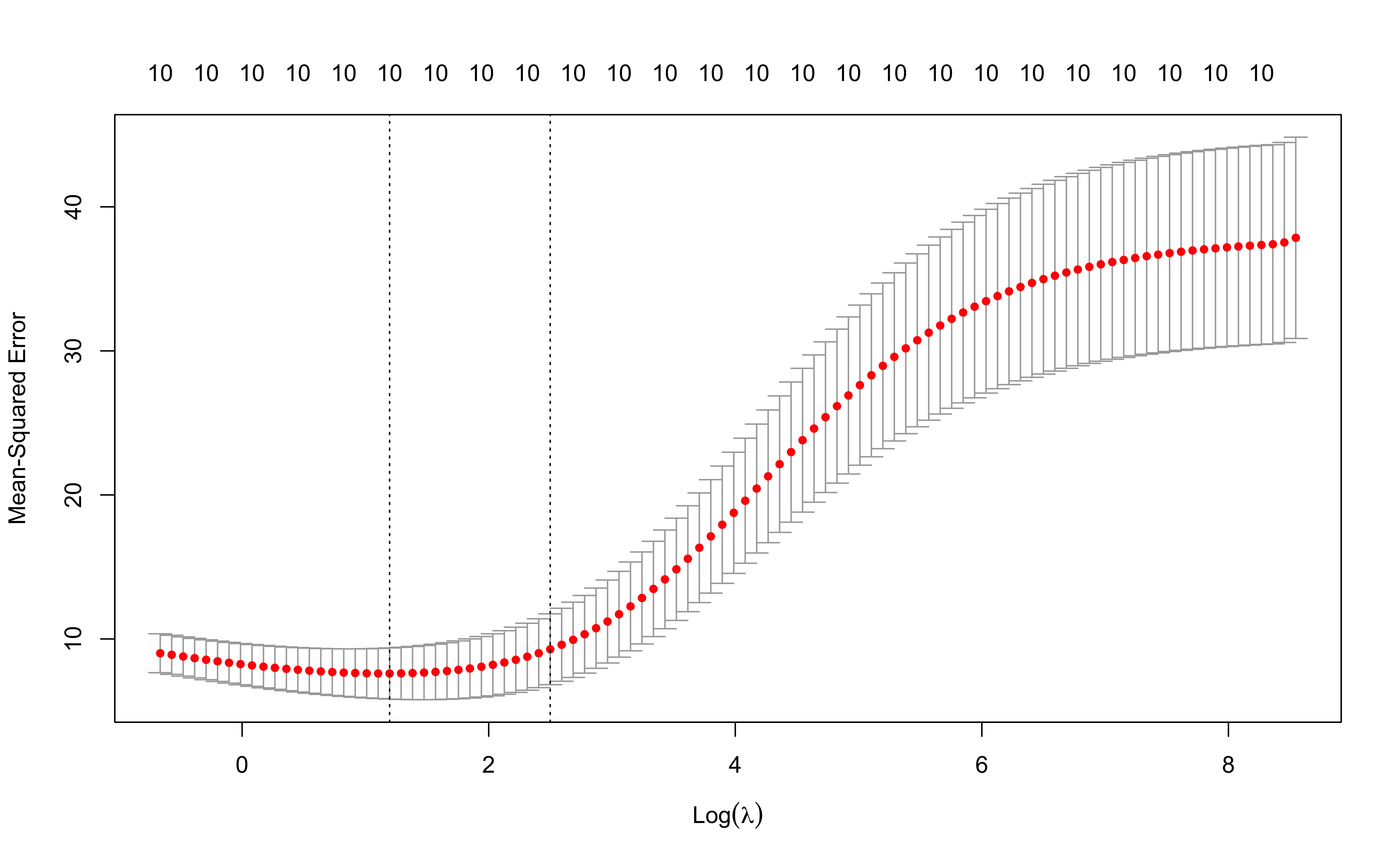
ridge_cv_fit$lambda.min[1] 3.31# largest lambda s.t. error is within 1 s.e of the min
ridge_cv_fit$lambda.1se [1] 12.2Generalized Cross-Validation
The generalized cross-validation (GCV) is a modified version of the leave-one-out CV (LOOCV) (\(n\)-fold CV).
The LOOCV for linear regression is
\[\text{CV}_{(n)} = \frac{1}{n}\sum_{i=1}^n \left[ \frac{y_i - x_i' {\mathbf{b}}} {1 - {\bf H}_{ii}} \right]^2 \]
- The GCV criterion is given by
\[\text{GCV}(\lambda) = \frac{1}{n}\sum_{i=1}^n \left[ \frac{y_i - x_i' \widehat{\boldsymbol \beta}^\text{r}_\lambda}{1 - \frac{\text{Trace}(\mathbf{S}_\lambda)}{n}} \right]^2\]
where \(\mathbf{S}_\lambda\) is the hat matrix corresponding to the ridge regression:
\[\mathbf{S}_\lambda = \mathbf{X}(\mathbf{X}' \mathbf{X}+ n\lambda \mathbf{I})^{-1} \mathbf{X}'\]
Generalized Cross-Validation
Select the best \(\lambda\) that produces the smallest GCV error.
ridge_fit <- lm.ridge(mpg ~ ., data = mtcars, lambda = 0:40)plot(ridge_fit$lambda,
ridge_fit$GCV,
type = "l", col = "darkgreen",
ylab = "GCV", xlab = "Lambda",
lwd = 3)